This is the second in a yearlong series of stories showcasing the research that the Ocean Protection Council supported in partnership with California Sea Grant, with funding from Proposition 84.
The warm El Niño currents drifting north toward Santa Barbara in 2015 brought exciting new visitors: the waters off the coast of the city suddenly featured wahoo and yellowtail — big, tropical fish that can make for a thrilling catch, but had previously been only caught further south in Mexico.
Other southern species were spotted, too, including a whale shark and several pufferfish. Warming oceans mean these out-of-place fish may be seen in the region more often. What’s not yet clear is whether these fish will become established in new territory. For that, fish will not just have to temporarily visit but also breed up north — a situation that’s harder to track since it can take years to observe if an adult population is reproducing and thriving.
“To understand the geographic distribution of species and how it’s influenced by climate change, we need to know more than just where the fish are,” says Ron Burton, a professor of marine biology at Scripps Institution of Oceanography at the University of California, San Diego. Using Ocean Protection Council funding, and supported by California Sea Grant, Burton set out to study where these fish are spawning.
For three years, from 2019 to 2021, Burton’s team sampled the water off five piers along the California coast for fish eggs. Then came the hard part — determining which species had spawned them. Though they can vary slightly in size, most fish eggs are transparent tiny spheres, about as big as poppy seeds. It's almost impossible to distinguish fish eggs based on their form and structure alone.
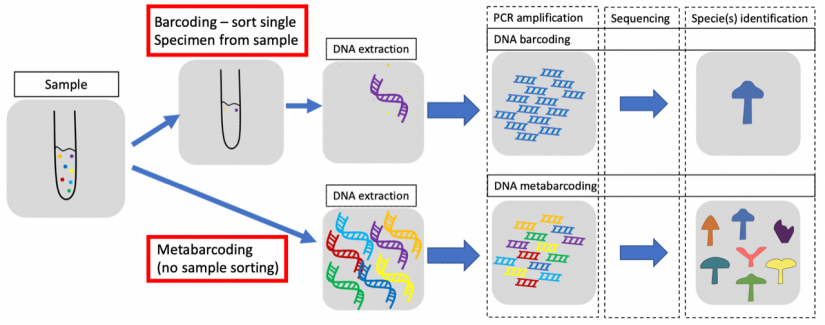
Over the last two decades, scientists have learned to identify fish eggs using their DNA. By extracting mitochondrial DNA, sequencing it and comparing specific sections with current databases, researchers can determine the species at hand. Since this approach treats the DNA as something akin to the barcodes in a grocery store, it became known as “barcoding.”
Identifying one egg through barcoding costs around $2.50, Burton notes. That might seem cheap, but a single zooplankton sample can contain as many as a thousand fish eggs, quickly escalating the price tag. Burton decided to try a new approach known as “metabarcoding” to see if it was a suitable alternative.
With metabarcoding, researchers do not extract DNA from individual eggs; instead, many eggs are ground up and their pooled DNA is sequenced together. The species’ different barcodes can still be read, though they can no longer be matched to specific eggs. Instead, scientists use this process to determine the rough proportions of the species within each sample: what percentage of eggs are from species A, what percentage are from species B, and so on. By using metabarcoding, a researcher sacrifices a bit of precision in exchange for less labor and thus potentially lower costs.
But found that many of the samples he collected contained twenty eggs or less. Metabarcoding costs around $50 per sample — meaning that for most of his team's specimens, it was still cheaper to use barcoding. “We’re now well into ten years of sampling from Scripps Pier and still using the same approach as when we started,” Burton says. He does not mean that as a bad thing. He notes that some researchers are eager to use whatever tool is newest, but sometimes the older tools still work best.
Burton also compared metabarcoding to a technique that analyzes what is known as “environmental DNA,” or, for short, eDNA. The term refers to bits of genetic material that have sloughed off biological cells and float in the water itself. For Burton’s purpose, eDNA has one notable disadvantage: there’s no way of knowing which DNA stems from eggs and larvae, and which from full-grown fish. Still, Burton found that he could learn something new from eDNA. By analyzing the same sample twice — using barcoding or metabarcoding for the eggs, then separately analyzing the eDNA — he can pinpoint which species are spawning and which are merely present without reproducing.
This technique also helped answer one of Burton's original questions: whether the newly arrived fish spurred northward by climate change have begun to spawn in their new range. For now, the answer seems to be no. Too few fish are making the move and many likely die when the waters grow cold in winter, or they’re simply failing to reproduce in their new location. Despite initial forays north, fish ranges don’t appear to have substantially shifted yet.
Things might look different in the future. “I feel like we got a nice baseline,” Burton says. “Hopefully, ten years from now somebody will be able to use it and see whether reproductive populations have shifted to the north.”
About California Sea Grant
NOAA’s California Sea Grant College Program funds marine research, education and outreach throughout California. Headquartered at Scripps Institution of Oceanography at the University of California San Diego, California Sea Grant is one of 34 Sea Grant programs in the National Oceanic and Atmospheric Administration (NOAA), U.S. Department of Commerce.